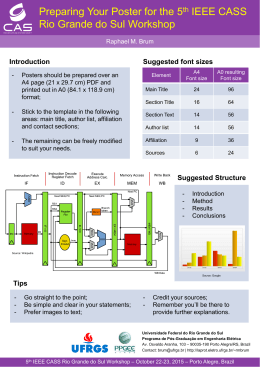
IDENTIFICATION OF POTENTIAL MIRNAS AND THEIR TARGETS IN VRIESEA CARINATA (POALES, BROMELIACEAE) Guzman, F1; Almerão, MP2; Korbes, AP1; Zanella, CM1; Bered, F1; Margis, R2* 1 PPGGBM at Federal University of Rio Grande do Sul - UFRGS, Porto Alegre, RS, Brazil. 2 Centre of Biotechnology and PPGBCM, Laboratory of Genomes and Plant Population, Federal University of Rio Grande do Sul - UFRGS, Porto Alegre, RS, Brazil. *E-mail: [email protected] Palavras-chave: Vriesea carinata; miRNAs; high-throughput sequencing; small RNAs; bioinformatics miRNAs play important functions in regulation of gene expression at the post-transcriptional level. A small RNA and RNA-seq of libraries were constructed to identify miRNAs in V. carinata, a native bromeliad species from Brazilian Atlantic Rainforest. Solexa technology was used to perform high throughput sequencing and data was analyzed using bioinformatics tools. We obtained 2,191,509 mature miRNAs representing 54 conserved families in plant species. Further analysis allowed the prediction of secondary structures of 19 conserved and 16 novel miRNAs. Potential targets were predicted from pre-miRNAs by sequence homology. This study provides the first identification of miRNAs and their potential targets of a bromeliad species without previous genomic sequence resources. Orgão financiador: PDJ, CNPq 8
Baixar