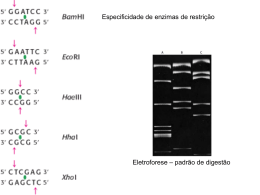
Seqüenciamento e Análise de Genomas 1 1953 – Modelo de Watson e Crick - O DNA é uma hélice dupla em que as bases nitrogenadas interagem por pontes de hidrogênio 2 1958 - A replicação do DNA é semiconservativa 3 Lewin, http://www.ergito.com Seqüenciamento de DNA Em 1977 o método de seqüenciamento com terminadores de cadeia é desenvolvido Sanger F, Nicklen, S and Coulson AR. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences (USA) 74: 5463– 5467, 1977 4 Seqüenciamento de DNA pelo método de Sanger 5 Seqüenciamento de DNA pelo método de Sanger 6 Manual sequencing uses radiolabeled dATP (35-S or 33-P) to label the DNA. The sample is then split into four tubes each with an individual ddNTP present. The samples are then subjected to acrylamide gel electrophoresis followed by autoradiography G A T C Manual Sequencing 7 O INÍCIO DA ANÁLISE DE GENOMAS 8 1977 – Sanger e colegas desenvolvem a técnica de seqüenciamento usando inibidores como terminadores de síntese de DNA. O primeiro genoma completo é seqüenciado - o do bacteriófago phi-x174 (5375 bases). Sanger F et al. The nucleotide sequence of bacteriophage phi-X174. Journal of Molecular Biology 125: 225-46, 1977. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chainterminating inhibitors. Proc Natl Acad Sci U S A. 74: 5463-5467, 1977 . 1980 – O método de Shotgun é desenvolvido Sanger e colaboradores desenvolveram o método de shotgun para preparar moldes para seqüenciamento de DNA: esse trabalho também apresentou o uso de vírus bacterianos (bacteriófago M13) para clonagem e preparação de moldes. Sanger et al. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. 1980. Journal of Molecular Biology 143 161-78 9 http://www.yourgenome.org/timeline.html Early genome sequencing projects The ability to undertake DNA sequencing on a large scale started a revolution in biology, as it became theoretically possible to determine the complete DNA sequence of any organism, and thus obtain a full description of the complete set of genes, or genome. Initially small genomes of viruses and organelles were studied (phiX174, 5.4kb; the mitochodrial genome, 16kb; bacteriophage lambda, 50kb; Epstein Barr virus, 172kb; human cytomegalovirus, 229kb). It became clear that the most efficient means to determine a complete genome sequence was to break the DNA randomly into fragments of appropriate size for sequencing, and then to reassemble the pieces using the DNA sequence itself. Overlaps between the individual pieces were identified on the basis of matches between independently sequenced DNA fragments. This process, termed random shotgun sequencing, was first applied to the whole genome of bacteriophage lambda, and has since been applied to progressively larger projects as the speed of sequencing and the power of computers to undertake the assembly process has increased. http://www.sanger.ac.uk/HGP/draft2000/early.shtml 10 1981 - IBM introduz o primeiro computador de uso pessoal – ele tinha 100 KB de memória, um driver de disco flexível e custava cerca de $3000 1982 - Compaq introduziu o primeiro computador portátil – sem disco rígido e custava cerca de $3000 1982 – O primeiro genoma é seqüenciado pela metodologia de shotugun de genoma total (whole genome shotgun WGS) Sanger e colaboradores seqüenciam o genoma do bacteriófafo Lambda usando a técnica de shotgun. Sanger F et al. 1982. Nucleotide sequence of bacteriophage lambda. Journal of Molecular Biology. 162: 729–73 11 Tamanho do genoma – 48.502 pb 1986 – O primeiro protótipo de uma máquina para seqüenciamento de DNA é apresentado. 1987 -Apple introduz o Mac II. Com um disco rígido de 40MB e custo de $5500 1987 - Olson e colegas desenvolvem o método para clonagem de longas regiões de DNA do genoma (100.000–200.000 pares de bases) Burke et al. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236 806-812, 1987. 12 1989 – O primeiro sistema de hipertexto – a base da world wide web – foi desenvolvido no European Organisation for Nuclear Research (CERN ) 1989 – Criação da Human Genome Organisation (HUGO) HUGO, uma associação internacional de pesquisadores envolvidos com o Projeto Genoma Humano foi criado para ajudar a coordenar as atividades e auxiliar na distribuição dos dados e recursos. 1990 – Apresentação, da Proposta formal do Department of Energy e National Institutes of Health, USA, para 13 seqüenciar o genoma humano. 1992 - Sulston e Waterston solicitam recursos da Wellcome Trust para seqüenciar 40Mb do genoma humano – proposta de cinco anos. Wellcome Trust e Medical Research Council, UK, associam-se ao Projeto Genoma Humano 1994 -Mosaic Communications – depois Netscape Communications – é criado 1995 – O primeiro genoma de um microrganismo de vida livre é seqüenciado O genoma de Haemophilus influenzae – 1.830.137 pares de bases – é seqüenciado pelo The Institute for Genomic Research (TIGR) 14 1997 - Seqüenciador automático MegaBace 1000 (96 capilares) 5 corridas/dia = ~200.000 pb 1998 – O seqüenciador ABI Prism 3700 é lançado, com capacidade para analisar oito conjuntos de 96 reações por dia =~308.000 pb 2001 - Seqüenciador automático MegaBace 4000 (384 capilares) Parte de um eletroferograma gerado no seqüenciamento 15 automático de DNA The MegaBACE 1000 DNA Sequencing System features: · Capillary electrophoresis with automated gel matrix replacement, sample injection, DNA separation, and base calling. Unlike slab-gel systems, the automation of gel replacement eliminates the need to pour gels, wash glass plates, and re-track samples after electrophoresis. · Electrokinetic sample injection automatically loads samples from a 96-well plate into the capillaries. Automated sample injection eliminates manual sample loading and sample mix up. · Energy transfer (ET) dye chemistry kits are the most robust and sensitive kits available. DYEnamic™ ET primers and DYEnamic ET terminator kits are formulated with Thermo Sequenase™ DNA polymerase and Thermo Sequenase II DNA polymerase, respectively, and are optimized for use with the MegaBACE. · The MegaBACE 1000 uses linear polyacrylamide (LPA) separation matrix. LPA is an innovation in capillary electrophoresis sieving matrices that enables read lengths in excess of 800 bases. With long read lengths; MegaBACE 1000 is ideal for finishing. · Flexible, user-friendly analysis software produces accurate base-calling data in a PHRED-compatible file format. 16 DYEnamic ET Terminators The dye terminators feature novel dye-labelled dideoxynucleotides and a new DNA polymerase. Each dideoxy terminator is labelled with two dyes. One of these dyes, fluorescein, has a large extinction coefficient at the wavelength (488 nm) of the argon-ion laser in the instrument. The fluorescein donor dye absorbs light energy from incident laser light and transfers the collected energy using radiationless energy transfer to an “acceptor” dye. Each of the four chain terminators, ddG, ddA, ddT, and ddC, has a different acceptor dye coupled with the fluorescein donor. The acceptor dyes then emit light at their characteristic wavelengths. The fluorescence is detected by the instrument, which allows the nucleotide that caused the termination event to be identified. Using energy transfer provides a more efficient excitation of the acceptor dyes than does using direct excitation by the laser, and results in a sequencing method that is very sensitive and robust. The acceptor dyes are the same standard rhodamine dyes used in DYEnamic ET primers: rhodamine 110, rhodamine-6-G, tetramethyl rhodamine, and rhodamine X. By using the standard rhodamine dyes as acceptors, the reaction products can be detected using the same filter set as the DYEnamic ET primers. The DYEnamic ET primers for the MegaBACE system are 5-carboxy-fluorescein (FAM) as the donor dye. The acceptor dyes are rhodamine 110 (R110) (C), 6carboxyrhodamine (REG) (A), N,N,N’,N’,-tetramethyl-5-carboxyrhodamine 17 (TAMRA) (G) and 5-carboxy-X-rhodamine (ROX) (T). Crescimento no número de seqüências no GenBank Year Base pairs Sequences 1982 680,338 606 1994 217,102,462 215,273 1995 384,939,485 555,694 1996 651,972,984 1,021,211 1997 1,160,300,687 1,765,847 1998 2,008,761,784 2,837,897 1999 3,841,163,011 4,864,570 2000 11,101,066,288 10,106,023 2001 15,849,921,438 14,976,310 2002 28,507,990,166 22,318,883 2003 36,553,368,485 30,968,418 2004 44,575,745,176 40,604,319 Revised: February 16, 2005. 18 Projetos Genoma Vários projetos de seqüenciamento de genomas bacterianos foram ou estão sendo desenvolvidos no mundo, enfocando as mais diversas espécies de organismos como Mycobacterium tuberculosis, Vibrio cholera e inúmeras outras bactérias (248 já concluídos e mais de 402 em andamento); Vários genomas eucarióticos também já foram seqüenciados. 19 Genome sequencing projects statistics Organism Prokaryotes Archaea Bacteria Eukaryotes Animals Mammals Birds Fishes Insects Flatworms Roundworms Amphibians Reptiles Other animals Plants Land plants Green Algae Fungi Ascomycetes Basidiomycetes Other fungi Protists Apicomplexans Kinetoplasts Other protists total: Complete 248 22 226 19 4 2 1 Draft assembly 127 3 124 54 20 7 1 2 6 1 2 2 2 2 9 7 1 1 4 2 2 267 25 21 3 1 9 5 1 3 181 In progress 275 11 264 131 61 18 2 25 2 3 1 12 19 15 4 20 16 2 2 27 9 3 14 406 total 650 36 614 204 85 27 1 4 32 2 6 1 0 14 21 17 4 54 44 6 4 40 14 6 19 854 Revised: Aug 16, 2005 http://www.ncbi.nlm.nih.gov/genomes/static/gpstat.html 20 Genomas eucarióticos human C. elegans Plasmodium falciparum Arabidopsis thaliana 21 Schizosaccaromyces pombe Drosophila melanogaster Whole Genomes The Draft Genome of Ciona intestinalis: Insights into Chordate and Vertebrate Origins Paramvir Dehal et al.Science, 13 December 2002, p. 2157-2167 The Genome Sequence of the Malaria Mosquito Anopheles gambiae Robert A. Holt et al. Science, 4 October 2002, p. 129-149 Whole-Genome Shotgun Assembly and Analysis of the Genome of Fugu rubripes Samuel Aparicio et al. Science, 23 August 2002, p. 1301-1310; published online July 25, 2002, 10.1126/science.1072104 A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. indica) Jun Yu et al. Science, 5 April 2002, p. 79-92 A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. japonica) Stephen A. Goff et al. Science, 5 April 2002, p. 92-100 Miniature Genome in the Marine Chordate Oikopleura dioica Hee-Chan Seo et al. Science, 21 December 2001, p. 2506 22 Genoma humano Nature 431: 931 (2004) 23 Estratégias para Seqüenciamento de Genomas Shotgun: o genoma é fragmentado ao acaso em pedaços pequenos (2,0 – 4,0 kb) que se sobrepõem. Esses fragmentos dão clonados num plamídeo (pUC18). Um número suficiente de clones é seqüenciado para gerar um conjunto de seqüências que corresponde a várias vezes o genoma (aproximadamente 10X). As seqüências (reads) são montadas com auxílio de programas de computador em regiões genômicas completas. Clone a clone: um conjunto de clones de cada cromossomo ou região do genoma é organizado, cada clone é seqüenciado e as seqüencias de cada clone são montadas num segmento maior. 24 Estratégias para Seqüenciamento Shotgun Clone a Clone 25 Cobertura do genoma C= N.r/G C= cobertura do genoma N= número de reads r= tamanho médio dos reads G= tamanho do genoma C= 8X N= número de reads r= 400 bp G= 1 Mb 10=N.400/106 N= 20.000 Genome coverage: average number of times a nucleotide is represented by a highquality base in random raw sequence. Full shotgun coverage: genome coverage in random raw sequence required to produce finished sequence, usually 8-10 fold. Partial shotgun coverage: typically 3-6X random coverage of a genome which produces sequence data of sufficient quality to enable gene identification but which is not sufficient to produce a finished genome sequence Contig: contiguous DNA sequence produced from joining overlapping raw sequence reads. Scaffold: a group of ordered and orientated contigs known to be physically linked to each other by paired read information. 26 Finished sequence: complete sequence of a genome with no gaps and an accuracy of > 99.9%. Estratégia de seqüenciamento shotgun DNA genômico Seleção por tamanho Reparacão das pontas Ligacão no vetor Biblioteca Seqüenciamento Montagem Contigs 27 Fechamento e anotação Estratégia para Fechamento PACE - PCR-Assisted Contig Extension 28 Carraro et al. BioTechniques 34:626-632 (March 2003) 29 Lab Bioinformática – LNCC 30 Dra. Ana Tereza Vasconcelos Diagrama mostrando as etapas de um projeto de seqüenciamento 31 de genoma por “shotgun”. Fraser et al. (Nature 406, 799 – 803, 2000) O genoma de H. inflluenzae tem 1,83 Mb e contém 1740 open reading frames (ORFs) O genoma de M. genitalium tem 580.074 bp e contém 470 ORFs 32
Baixar