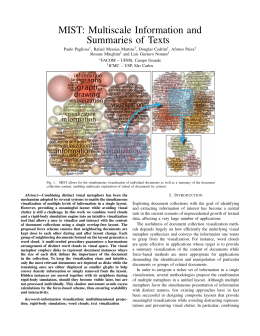
XLIII Annual Meeting of SBBq th th Foz do Iguaçu, PR, Brazil, May 17 to 20 , 2014 Visualization of Protein Folding Funnels in Structure-based Models Oliveira Junior, A. B.; Leite, V. B. P. Departamento de Física, IBILCE, UNESP, São José do Rio Preto, SP, Brazil Introduction: Protein folding occurs in a very high dimensional phase space, whose exponentially large number of states, represented in terms of one effective reaction coordinate, exhibit a topology resembling a funnel, according to the energy landscape theory. Since the role of each local minimum is not considered in this statistical approach, the folding mechanism is unveiled by describing the local minima in an effective one-dimensional representation. Objectives: Apply a metric to describe the distance between any two conformations, which allows to go beyond the one-dimensional representation and visualizing the folding funnel in 2D and 3D. Material and Methods: Computer simulation of protein folding were performed using structure-based model, particularly the C-alpha approximation. Two distinct proteins were studied, CI2 and SH3 (from the 1YPA and 1SHG pdb, respectively). The projection of these multidimensional data was performed using a metric based on the ratio between conformational similarity (Jaccard index) and dissimilarity (Jaccard distance). Results and Discussion: The visualization allows assessing the folding process in detail, e.g. by identifying the connectivity between conformations and establishing the paths that lead to the native state. Distinct funnels were generated according to the native state structure. The analysis was restricted to conformations from the transition-state to the native configuration. Due to the large number of structures, the visualization was divided into regions of energy, allowing better description of the states close to the native state. Conclusion: Consistent with the expected results from the energy landscape theory, folding routes can be visualized to probe different regions of the phase space, as well as to characterize metastable states. This methodology can be applied to infer the conformational effects of small mutations. Keywords: energy landscape theory, multidimensional reduction, protein comparison metric Acknowledgments: CAPES, FAPESP, CNPq
Download